Genomic Inbreeding
Genomic inbreeding is a powerful and accurate way to assess animal inbreeding.
Introduction
As of December 2022, Genomic inbreeding is calculated as part of AHDB’s genomic evaluation service, and available in the Herd Genetic Report for all official genomically tested cows. Inbreeding is increasingly being discussed and genomic analyses offer a more accurate and biologically meaningful estimate of an individual’s own inbreeding, as well as a guide for inbreeding of future matings. Monitoring inbreeding is recommended to managed inbreeding, while maximising genetic progress. Mating tools such as genomic inbreeding calculators help ensure that the beneficial effect of genetic progress continues to outpace any detrimental effect of inbreeding.
Summary
If you are genomic testing:
- Genomic inbreeding is now calculated as part of AHDB’s genomic evaluation services. For females it can be accessed in the Herd Genetic Report and is also available for checking future matings in the inbreeding checker.
- Genomic inbreeding calculations are superior to pedigree inbreeding estimates, especially when traditional pedigree records are partial or inaccurate.
- Genomic inbreeding coefficients scale similarly to pedigree inbreeding coefficients, so the same thresholds and breeding decisions can be applied to both.
What is inbreeding?
Inbreeding refers to the mating of related individuals (where the sire and the dam share a recent common ancestor). In pure-bred mating programmes inbreeding is unavoidable, however, not all inbreeding has a negative impact. Selection of beneficial traits such as production, health, and fertility can also increase inbreeding levels, while having a net positive effect on genetic progress. Nonetheless, it is important to monitor inbreeding and the potential negative effects that can accumulate. This can be achieved through management decisions such as, genotyping for specific disease-causing recessive mutations, using PTAs and indexes to counteract negative traits (such as Fertility and mastitis), or more broadly by checking inbreeding coefficients (preferably genomic inbreeding coefficients).
Pedigree inbreeding
Using pedigree information is the traditional approach to estimating inbreeding. This involves counting the number of common ancestors that occur in the back pedigree and weighting their contributions to inbreeding, based on which generation these common ancestors occur. This can be quantified into an inbreeding coefficient, given as a percentage between 0-100%. Table 1 shows inbreeding coefficients calculated on pedigrees that share a single notable common ancestor, however, real pedigrees are often more complicated and involve multiple common ancestors across generations.
Table 1. Pedigree inbreeding coefficient estimates.
| Relationship | Pedigree Inbreeding Coefficient |
| Animal mated to its own parent (e.g. Sire-Daughter) |
25% |
| Full sibling mating (parents share both sire AND dam) |
25% |
| Half sibling mating (parents share a sire OR dam) |
12.5% |
| Animal has a single common great grand parent |
3.1% |
Disadvantages of pedigree inbreeding estimates
While pedigree-based inbreeding coefficients are useful as a first step of estimating inbreeding, and have served us well, there are notable shortcomings:
- Pedigrees are not always complete – missing pedigree entries (e.g., no dam pedigree) can wipe out huge portions of the ancestral tree when attempting to calculate pedigree-based inbreeding coefficients, this can lead to underestimation of inbreeding, potentially leading to misinformed decisions.
- Pedigrees can be unreliable – an incorrect pedigree (e.g., wrong sire) can be more of an issue than an incomplete pedigree as a poor mating decision may be made with (misplaced) confidence.
- Pedigree estimates are NOT a direct measure of inbreeding – as each individual has two copies of each gene and only one copy (per parent) is passed to the offspring, it is impossible to be certain which copy has been passed to offspring using only pedigree data. We are familiar with the fact that two full sisters can be different in performance as a result, but the same underlying genetics also impact their inbreeding. Scaled across multiple generations, this can have a large impact on our ability to predict the true inbreeding of individuals and their offspring.
Genomic inbreeding and advantages
The passing of genetic information from one generation to another has an element of randomness to it, two siblings that share a sire and dam may have inherited completely different versions of genes from each parent. The accumulation of this effect across several generations can result in two individuals that share a similar pedigree to be genetically very different. Genomic inbreeding coefficients measure the proportion of the individual’s DNA that has been inherited from recent common ancestors, and in doing so gives a measure of the percent of the individual’s genome that is inbred.
Using genomic data to calculate inbreeding is a powerful alternative to pedigree-based estimates, with two significant advantages:
- Genomic inbreeding is not affected by shallow or incomplete pedigrees – genomic inbreeding coefficients are calculated independently to pedigree information, meaning that accurate estimates are calculated even if the pedigree is wrong or completely missing.
- Genomic estimates are a direct measure of inbreeding – genomic inbreeding looks directly at the variation present in an individual’s genome and measures the proportion of the genome inherited from recent common ancestors, giving a direct estimate of inbreeding.
Accessing genomic inbreeding
Currently the genomic inbreeding data that is available is:
- Female individual genomic inbreeding – Check how inbred the females in your herd are. This is available through the Herd Genetic Report under “Inbreeding Coefficient” in youngstock and milking herd reports. Genomic values can be distinguished from pedigree estimates by the genomic flag
 .
. - Inbreeding checker mating predictions – Use predictions of potential matings to make breeding decisions. This is available through the Herd Genetic Report under “Inbreeding Checker”. Predictions for the genomic inbreeding of offspring generated for mating sires and dams are available when the sire and dam have both been genomically tested.
The effect of inbreeding
Inbreeding has a measurable effect on an animal’s phenotype, including production, fertility, health, and survival. When an inbreeding event occurs, both negative and positive traits can become fixed within an individual; the selection process aims to purge the negative effects while retaining the positive effects, however, this can take multiple generations. This results in recent inbreeding having a stronger negative influence than ancient inbreeding.
Our preliminary results show that every 1% increase in inbreeding is associated with a ~0.35% loss in production (milk, fat, and protein) over a cow’s lifetime, with the majority of this linked to recent inbreeding. There is also an indication that ancient inbreeding can have a net positive influence on traits. Similar trends are observed in both Canadian (Bayode et al. 2020) and Dutch (Doekes et al. 2019) Holstein.
FAQs
Q: How much will the genomic inbreeding value differ from pedigree-based inbreeding estimates?
A: Genomic inbreeding is on the same scale as pedigree-based inbreeding, ranging from 0-100%. There will be slight differences in genomic and pedigree estimates, however, when comparing genomic inbreeding with pedigree inbreeding for animals that have extensive, high-quality pedigrees there is over a 90% correlation in the values. The difference is greater when pedigree estimates were previously estimated from poor or incorrect pedigrees.
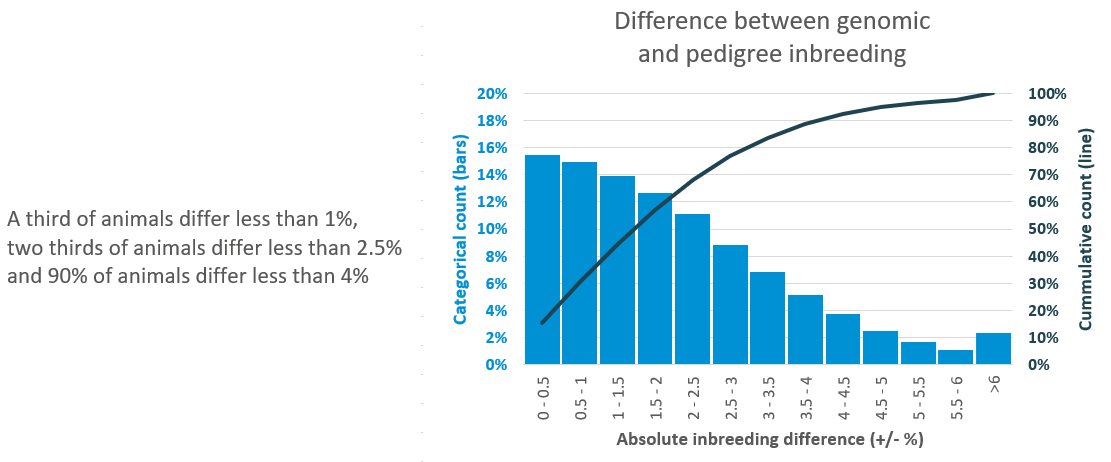
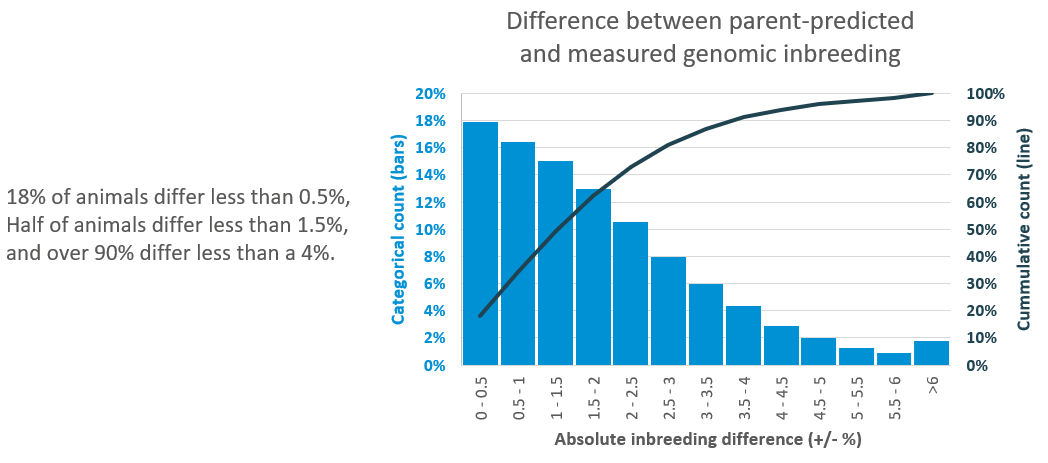
Q: Why is there a difference between the inbreeding checker genomic mating estimates and the genomic individual inbreeding for offspring born from that mating?
A: There is an element of randomness in the inheritance of DNA from parents, in the same way that all full siblings are not identical. The inbreeding checker gives the average expected inbreeding of potential offspring, however, naturally there is some variation around this value.
Q: How will I know it’s a genomic inbreeding value and not a pedigree-based estimate?
A: Genomic inbreeding values will be flagged with ![]() , if an animal in the Herd Genetic Report is flagged as genomic, then the inbreeding coefficient will be a genomic estimate. For the inbreeding checker, each mate pairing that is a genomic estimate will be flagged with
, if an animal in the Herd Genetic Report is flagged as genomic, then the inbreeding coefficient will be a genomic estimate. For the inbreeding checker, each mate pairing that is a genomic estimate will be flagged with ![]() .
.

