- Home
- Research Bite Two: Can we predict lameness in dairy cows using metabolomics?
Research Bite Two: Can we predict lameness in dairy cows using metabolomics?
Metabolomics is a relatively young scientific study area of chemical processes involving metabolites, the end products of cellular interactions. It has been successfully used for predicting and diagnosing diseases or understanding their pathogenesis in humans. Ana Ferreirais part of a UK team researching the potential of using this science to predict lameness in dairy cows. Here, she explains a little about their research.
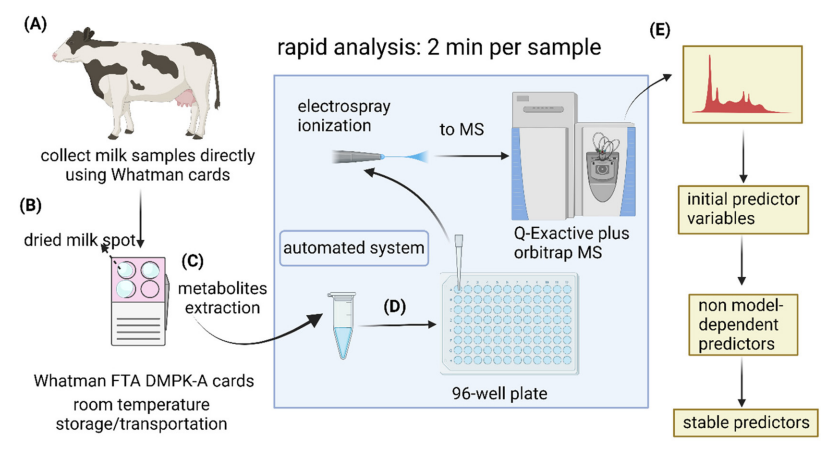
As in humans, the use of metabolomics in livestock has great potential for applications including the diagnosis or prediction of conditions such as lameness, particularly if predictive biomarkers can be identified that are detectable prior to changes in gait. To investigate whether this approach could be used to diagnose lameness in dairy cows, a new workflow (Figure 1) for untargeted metabolomics using direct infusion tandem mass spectrometry to analyse dry milk spots (DMS) was developed in work supported by the Academy of Medical Sciences and a BBSRC PhD studentship. Milk, being the most convenient biofluid sample to collect on dairy farms, was collected using cards (Whatman® FTA® DMPK) that are more commonly used to collect dried blood spots for diagnostic testing in humans. These cards are air-dried and can be stored at room temperature, adding to the convenience of sample collection prior to analysis.
 Ana Ferreira
Ana Ferreira
Figure 1. Workflow developed for collection of milk samples and analysis using mass-spectrometry
The milk drops were collected from 21 cows (10 lame and 11 healthy cows) from a dairy herd of 300 cows at the Center for Dairy Science Innovation (CDSI) at the University of Nottingham. The AHDB mobility scoring 0–3 scale was used, with cows greater than or equal to 2 being classified as lame and those with a mobility score of less than 2 classified as non-lame.
Metabolite extraction was preceded by removing part of each spot from the cards using a hole puncher on day eight. The extraction procedure was adapted from a method using dried blood spots. Then, solvents containing extracted metabolites were directly infused into a high-resolution mass spectrometer for a rapid metabolomic analysis via chip-based nanoelectrospray ionisation.
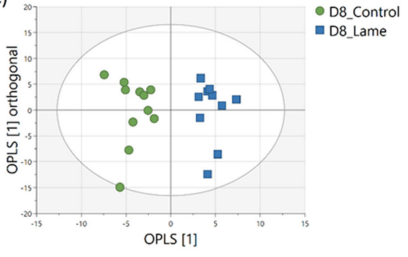
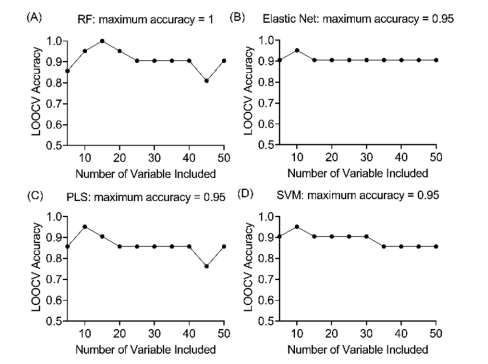
Important predictor metabolites (a set of lipid molecules and several small metabolites) were discovered by comparing the metabolome profiles from lame and control cows after orthogonal partial least squares-discriminant analysis (OPLS-DA; Figure 2). This was validated by triangulation of multiple machine learning models (Elastic net, Partial Least Squares and Support Vector Machine reached the highest accuracies of 95.2%; Figure 3). Stability selection was employed to reliably identify the most important discriminative metabolites. The addition of machine learning and stability selection to the conventional workflow was intended to mitigate the issue of method-dependent outcomes and identify the most stable predictors of lameness. To confirm the identities of the metabolites, both experimental and computed MS/MS spectra were used for structure-based identification.
 Ana Ferreira
Ana Ferreira
Figure 2. Results of comparison of mass-spectral data using OPLS of milk samples from lame versus sound cows
 Ana Ferreira
Ana Ferreira
Figure 3. Results of analysis of mass-spectral data using machine learning models to predict lameness
In summary, these findings met the expectations of the authors to prove the viability of using the easy collection of milk samples to identify metabolites that are highly predictive of lameness in a group of dairy cows. Validation work is now required to demonstrate the repeatability of results on another group of cows. If successful, this rapid, robust and discriminatory method shows potential to be used as a convenient and cost-effective sampling procedure for large-scale research and perhaps for the future routine diagnosis of lameness.
Editor’s note: Our understanding is that there is still some way to go before this might be commercialised and available for use in practice. However, the potential advantage for assisting early detection for EDPET is immense. We watch this space with fingers crossed! (Owen Atkinson)

